Make a classification model and run evaluations.
Part A
We are going to use a toy dataset called bivariate. There is a training, testing, and validation dataset provided.
library(tidyverse)
## Warning: package 'tidyverse' was built under R version 4.1.3
## -- Attaching packages --------------------------------------- tidyverse 1.3.2 --
## v ggplot2 3.4.0 v purrr 0.3.5
## v tibble 3.1.8 v dplyr 1.0.10
## v tidyr 1.2.1 v stringr 1.5.0
## v readr 2.1.3 v forcats 0.5.2
## Warning: package 'ggplot2' was built under R version 4.1.3
## Warning: package 'tibble' was built under R version 4.1.3
## Warning: package 'tidyr' was built under R version 4.1.3
## Warning: package 'readr' was built under R version 4.1.3
## Warning: package 'purrr' was built under R version 4.1.3
## Warning: package 'dplyr' was built under R version 4.1.3
## Warning: package 'stringr' was built under R version 4.1.3
## Warning: package 'forcats' was built under R version 4.1.3
## -- Conflicts ------------------------------------------ tidyverse_conflicts() --
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()
library(tidymodels)
## Warning: package 'tidymodels' was built under R version 4.1.3
## -- Attaching packages -------------------------------------- tidymodels 1.0.0 --
## v broom 1.0.1 v rsample 1.1.1
## v dials 1.1.0 v tune 1.0.1
## v infer 1.0.4 v workflows 1.1.2
## v modeldata 1.0.1 v workflowsets 1.0.0
## v parsnip 1.0.3 v yardstick 1.1.0
## v recipes 1.0.3
## Warning: package 'broom' was built under R version 4.1.3
## Warning: package 'dials' was built under R version 4.1.3
## Warning: package 'scales' was built under R version 4.1.3
## Warning: package 'infer' was built under R version 4.1.3
## Warning: package 'modeldata' was built under R version 4.1.3
## Warning: package 'parsnip' was built under R version 4.1.3
## Warning: package 'recipes' was built under R version 4.1.3
## Warning: package 'rsample' was built under R version 4.1.3
## Warning: package 'tune' was built under R version 4.1.3
## Warning: package 'workflows' was built under R version 4.1.3
## Warning: package 'workflowsets' was built under R version 4.1.3
## Warning: package 'yardstick' was built under R version 4.1.3
## -- Conflicts ----------------------------------------- tidymodels_conflicts() --
## x scales::discard() masks purrr::discard()
## x dplyr::filter() masks stats::filter()
## x recipes::fixed() masks stringr::fixed()
## x dplyr::lag() masks stats::lag()
## x yardstick::spec() masks readr::spec()
## x recipes::step() masks stats::step()
## * Use suppressPackageStartupMessages() to eliminate package startup messages
theme_set(theme_bw())
data(bivariate)
ggplot(bivariate_train, aes(x=A, y=B, color=Class)) +
geom_point(alpha=.3)
Use logistic_reg and glm to make a classification model of Class ~ A * B. Then use tidy and glance to see some summary information on our model. Anything stand out to you?
log_model <- logistic_reg() %>%
set_engine('glm') %>%
set_mode('classification') %>%
fit(Class ~ A*B,
data = bivariate_train)
log_model %>% tidy()
## # A tibble: 4 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0.115 0.404 0.284 7.76e- 1
## 2 A 0.00433 0.000434 9.97 2.01e-23
## 3 B -0.0553 0.00633 -8.74 2.32e-18
## 4 A:B -0.0000101 0.00000222 -4.56 5.04e- 6
log_model %>% broom::glance()
## # A tibble: 1 x 8
## null.deviance df.null logLik AIC BIC deviance df.residual nobs
## <dbl> <int> <dbl> <dbl> <dbl> <dbl> <int> <int>
## 1 1329. 1008 -549. 1106. 1126. 1098. 1005 1009Part B
Use augment to get predictions. Look at the predictions.
test_preds <- log_model %>% augment(bivariate_test)
test_preds
## # A tibble: 710 x 6
## A B Class .pred_class .pred_One .pred_Two
## <dbl> <dbl> <fct> <fct> <dbl> <dbl>
## 1 742. 68.8 One One 0.730 0.270
## 2 709. 50.4 Two Two 0.491 0.509
## 3 1006. 89.9 One One 0.805 0.195
## 4 1983. 112. Two Two 0.431 0.569
## 5 1698. 81.0 Two Two 0.169 0.831
## 6 948. 98.9 One One 0.900 0.0996
## 7 751. 54.8 One One 0.521 0.479
## 8 1254. 72.2 Two Two 0.347 0.653
## 9 4243. 136. One Two 0.00568 0.994
## 10 713. 88.2 One One 0.910 0.0898
## # ... with 700 more rowsPart C
Visually inspect the predictions using the code below
# log_model, your parnsip model
# bivariate_train / bivariate_val, data from bivariate
# to plot the countour we need to create a grid of points and get the model prediction at each point
x_grid <-
expand.grid(A = seq(min(bivariate_train$A), max(bivariate_train$A), length.out = 100),
B = seq(min(bivariate_train$B), max(bivariate_train$B), length.out = 100))
x_grid_preds <- log_model %>% augment(x_grid)
# plot predictions from grid as countour and validation data on plot
ggplot(x_grid_preds, aes(x = A, y = B)) +
geom_contour(aes(z = .pred_One), breaks = .5, col = "black") +
geom_point(data = bivariate_val, aes(col = Class), alpha = 0.3)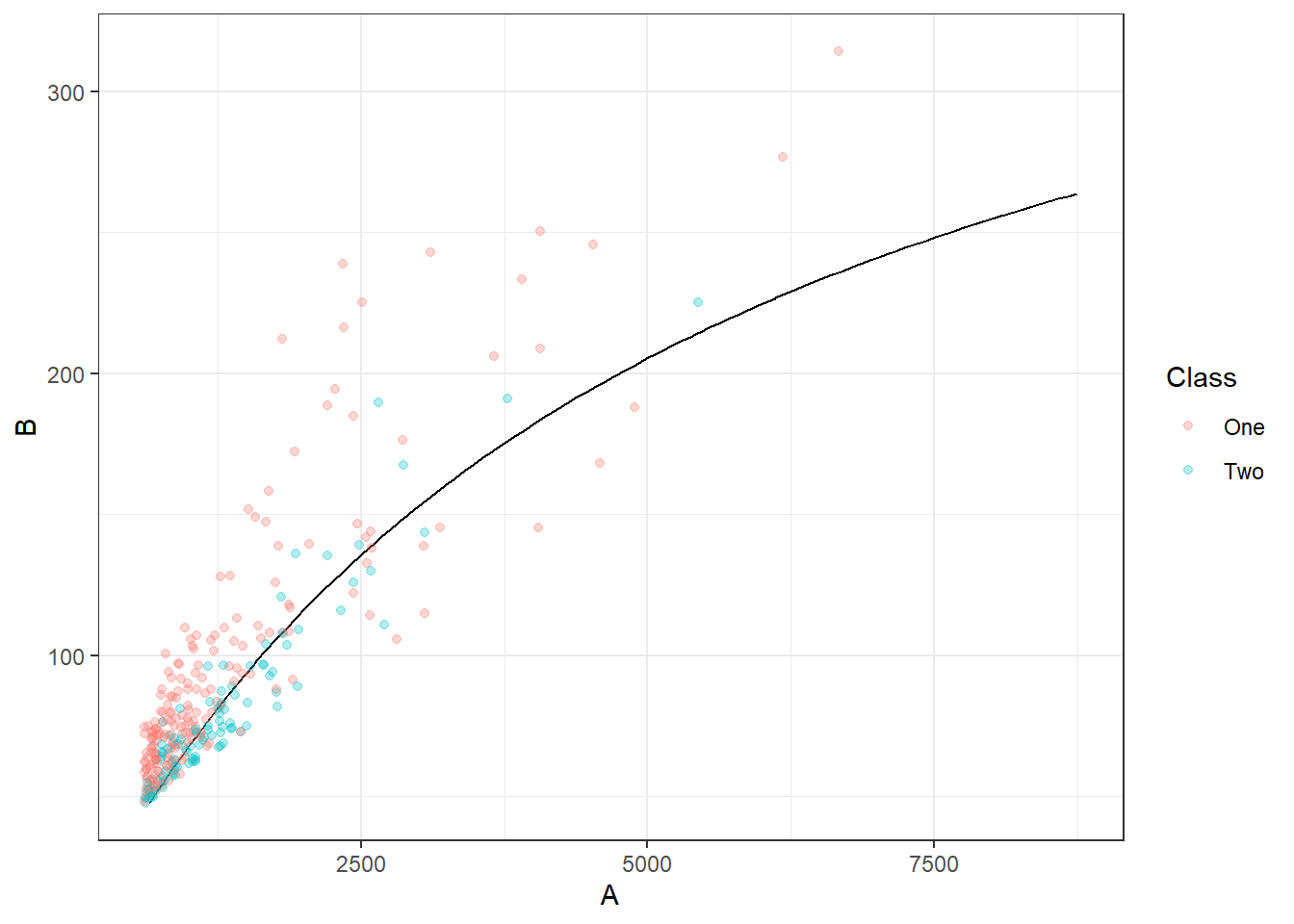
Part D
Evaluate your model using the following functions (which dataset(s) should you use to do this train, test, or validation). See if you can provide a basic interpretation of the measures.
- roc_auc
- accuracy
- roc_curve and autoplot
- f_meas
val_preds <- log_model %>% augment(bivariate_val)
val_preds %>% roc_auc(truth = Class,
estimate = .pred_One)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 roc_auc binary 0.790
val_preds %>% accuracy(truth = Class,
estimate = .pred_class)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.76
roc_curve(val_preds,
truth = Class,
estimate = .pred_One) %>%
autoplot()
f_meas(val_preds,
truth = Class,
estimate = .pred_class)
## # A tibble: 1 x 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 f_meas binary 0.827Part E
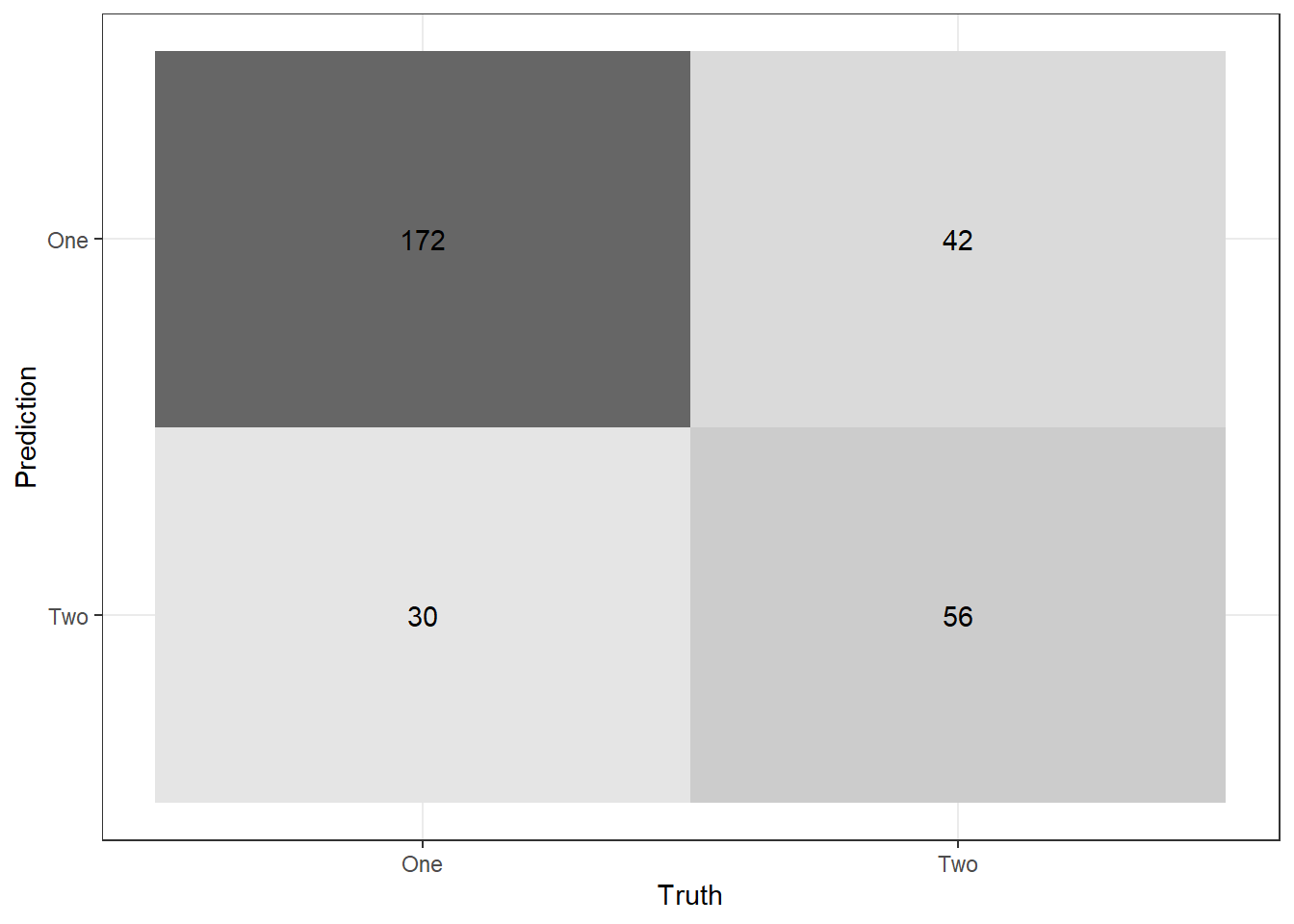
Recall Table 8.4 from the textbook. If necessary, class one can be positive and class two can be negative. Using the output from conf_mat, visually verify you know how to calculate the following:
- True Positive Rate (TPR), Sensitivity, or Recall
- True Negative Rate (TNR) or Specificity
- False Positive Rate, Type I error
- False Negative Rate (FNR), Type II error
- Positive Predictive Value (PPV) or Precision
val_preds %>% conf_mat(truth = Class,
estimate = .pred_class) %>%
autoplot("heatmap")